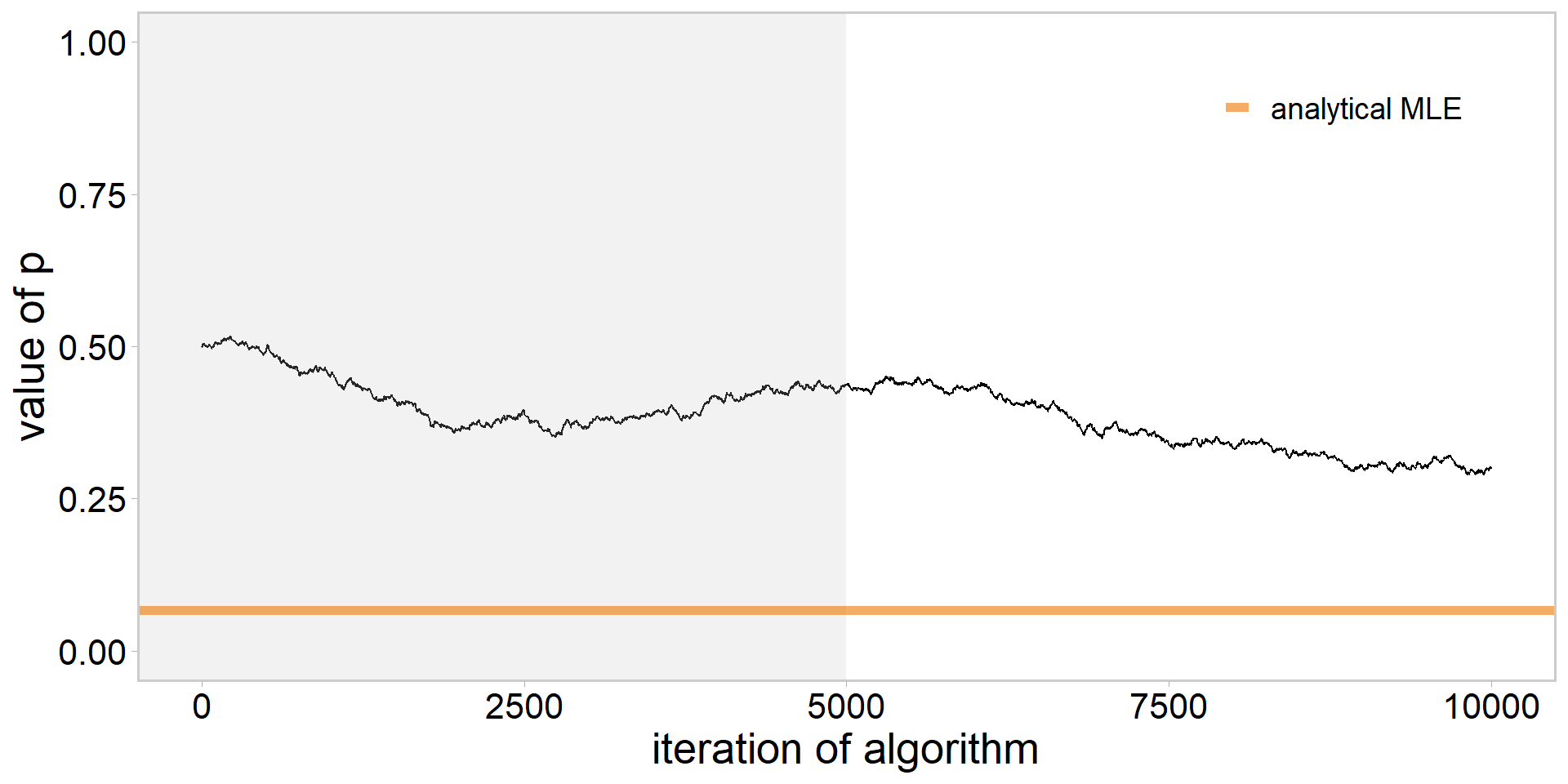
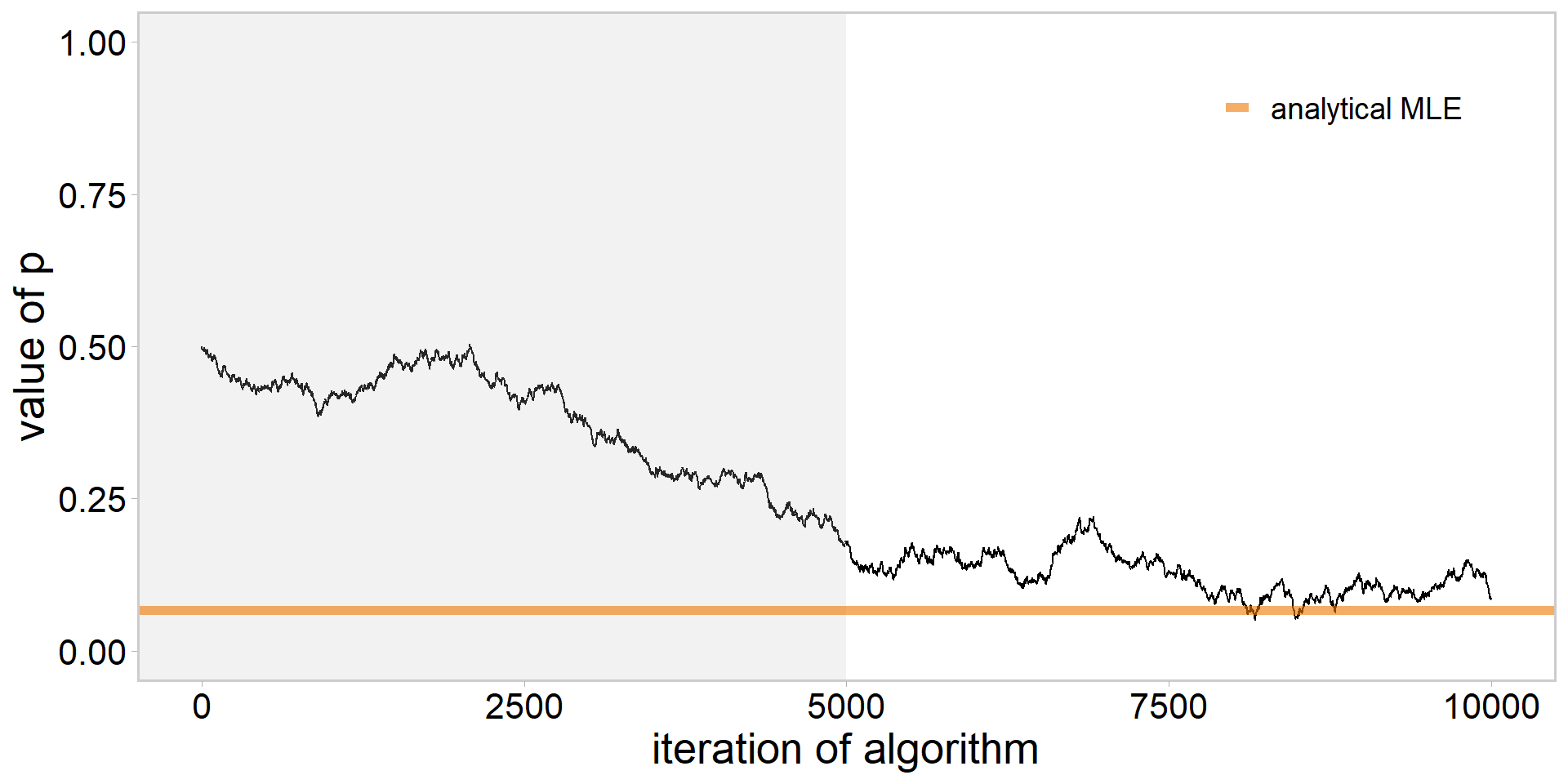
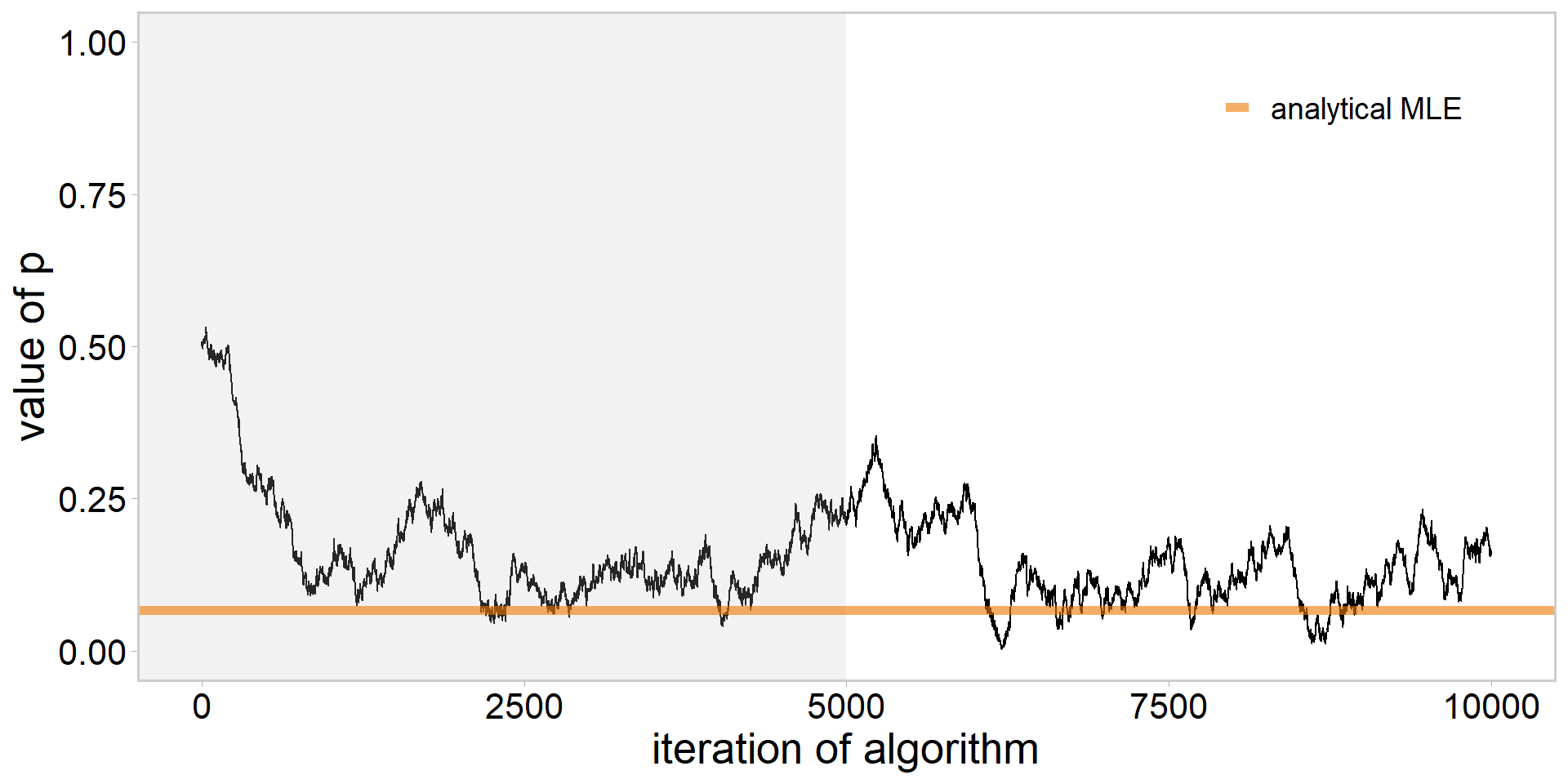
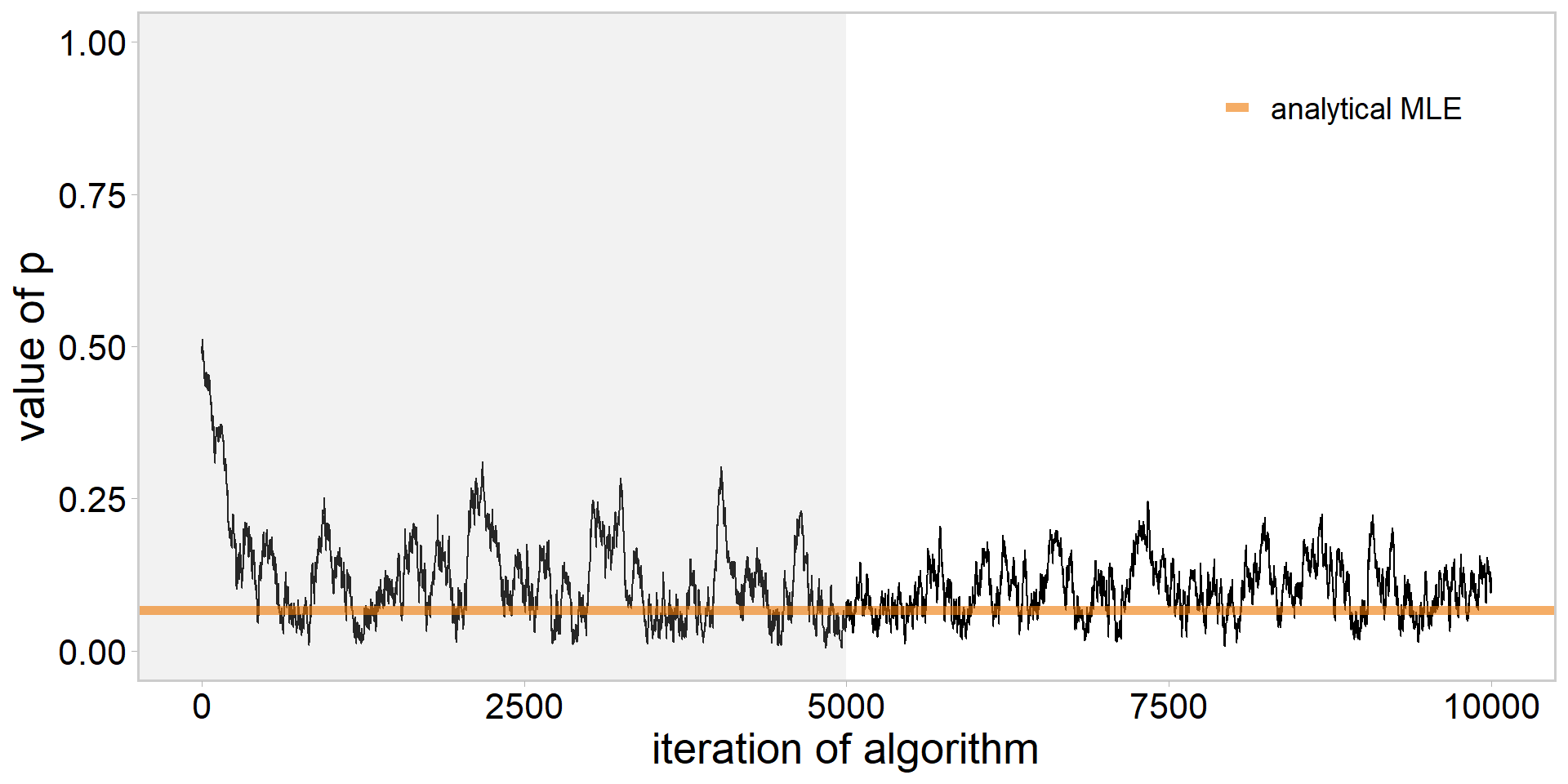
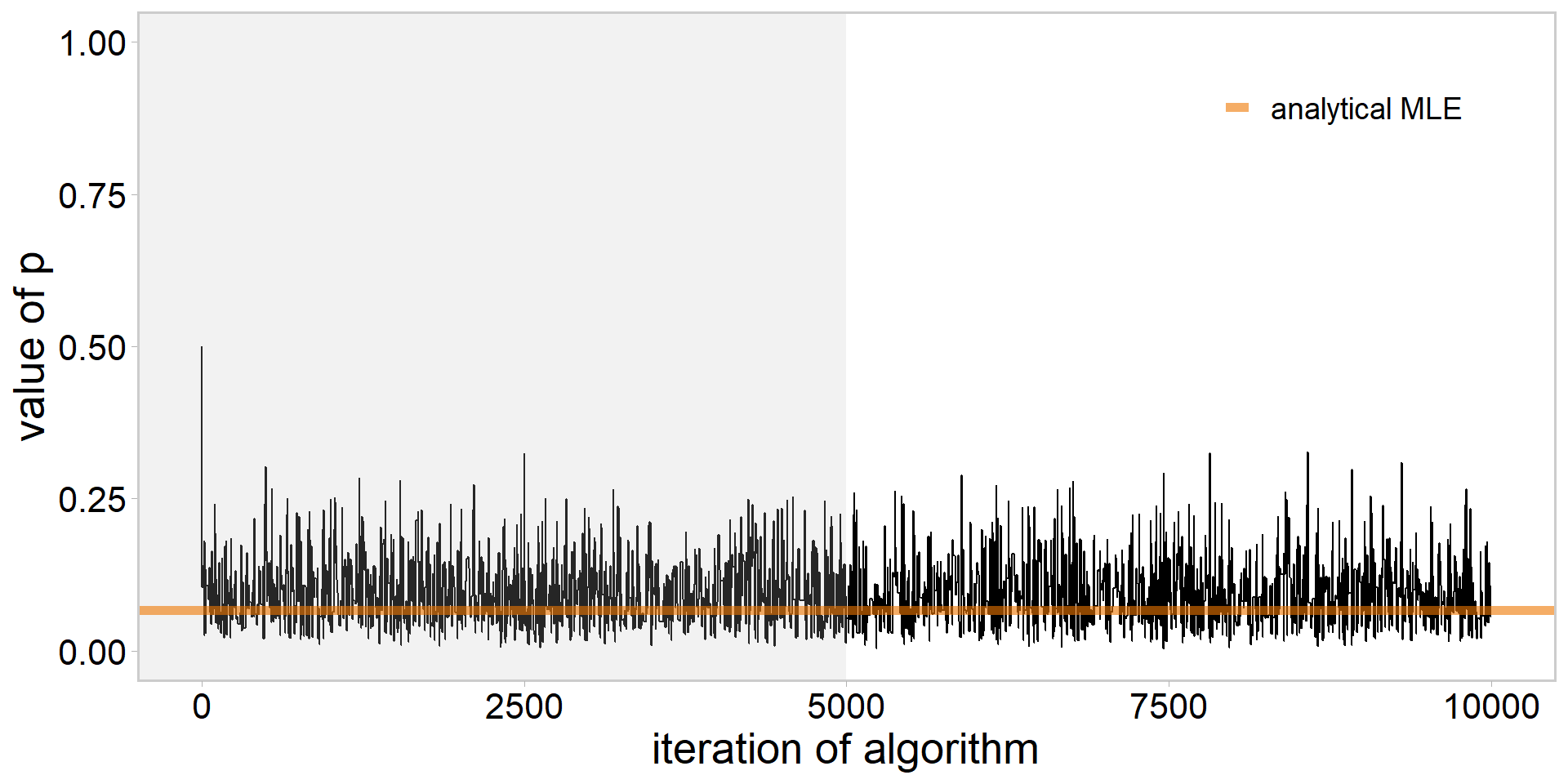
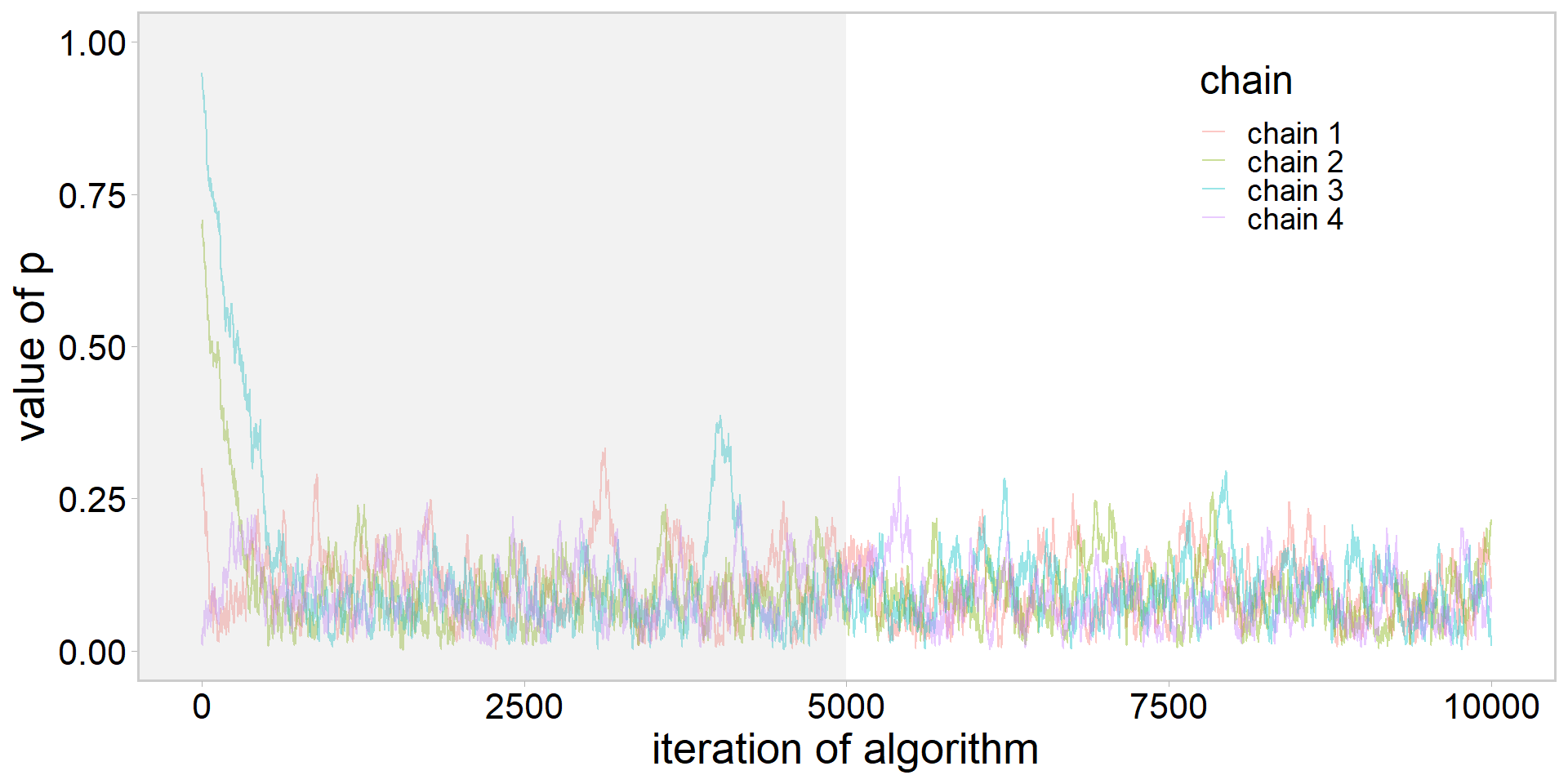
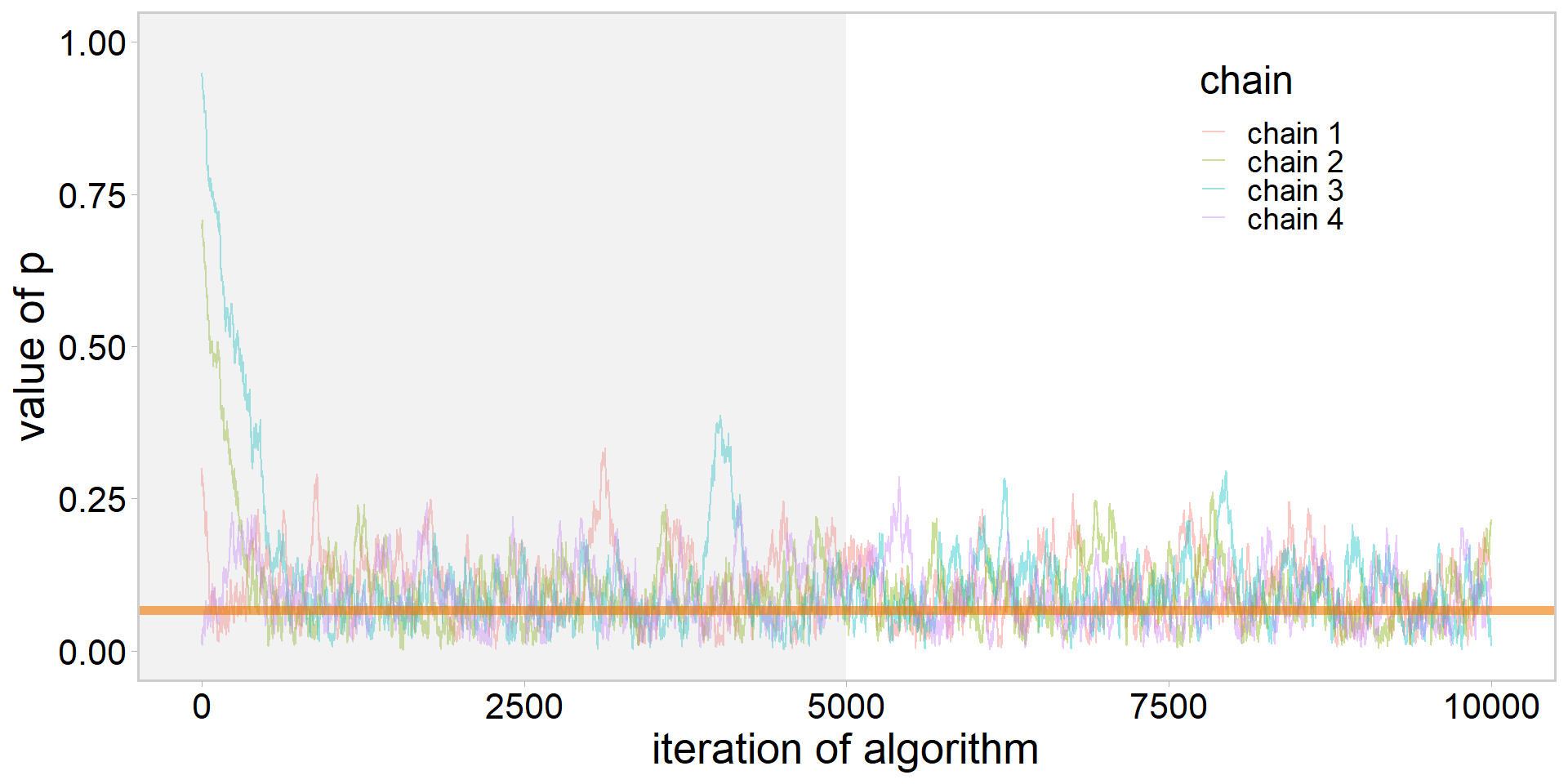
library(tidyverse)
library(ggqfc)
y <- c(
0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0
) # sucesses (turtle detections)
n <- length(y) # trials
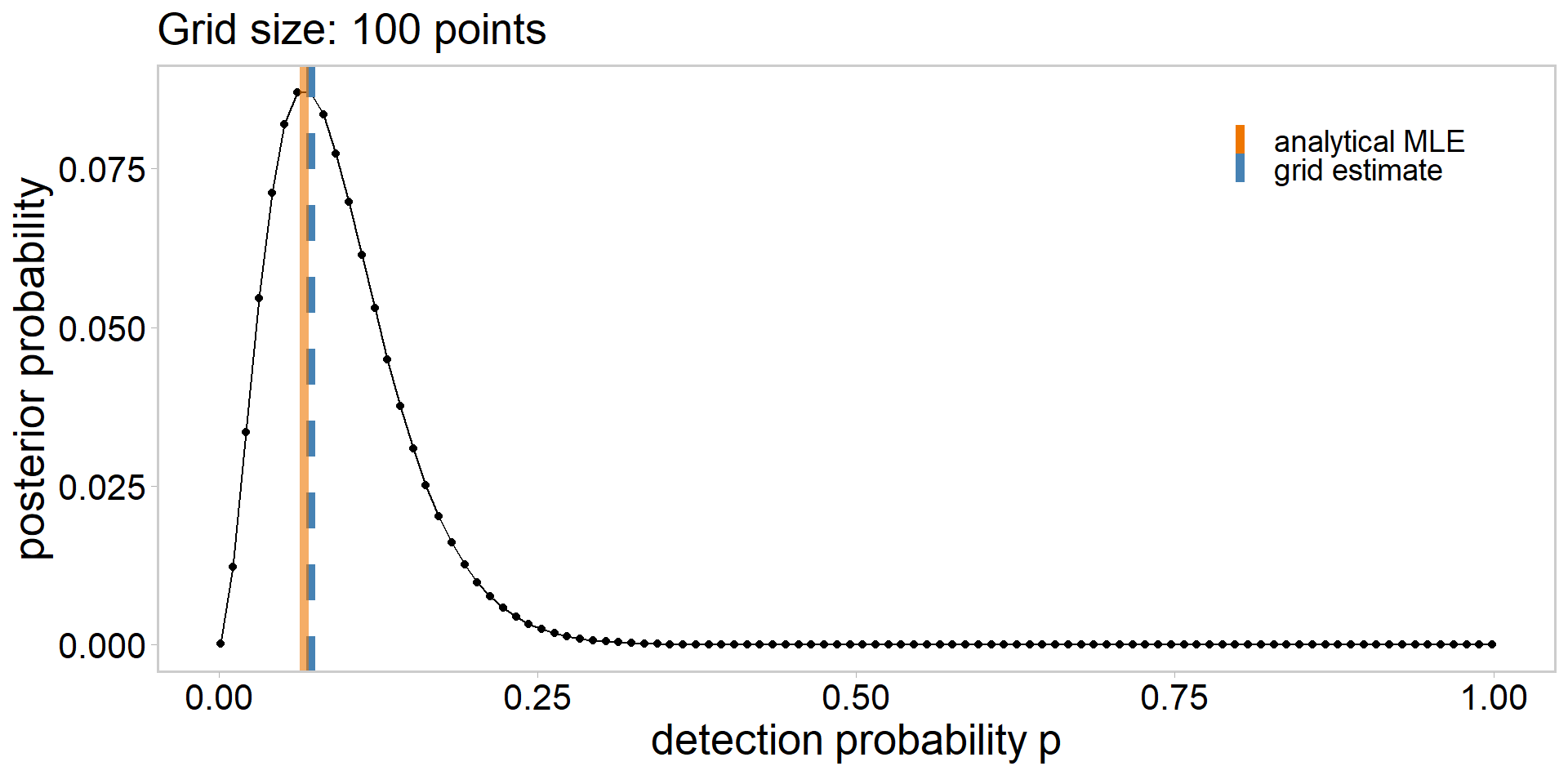
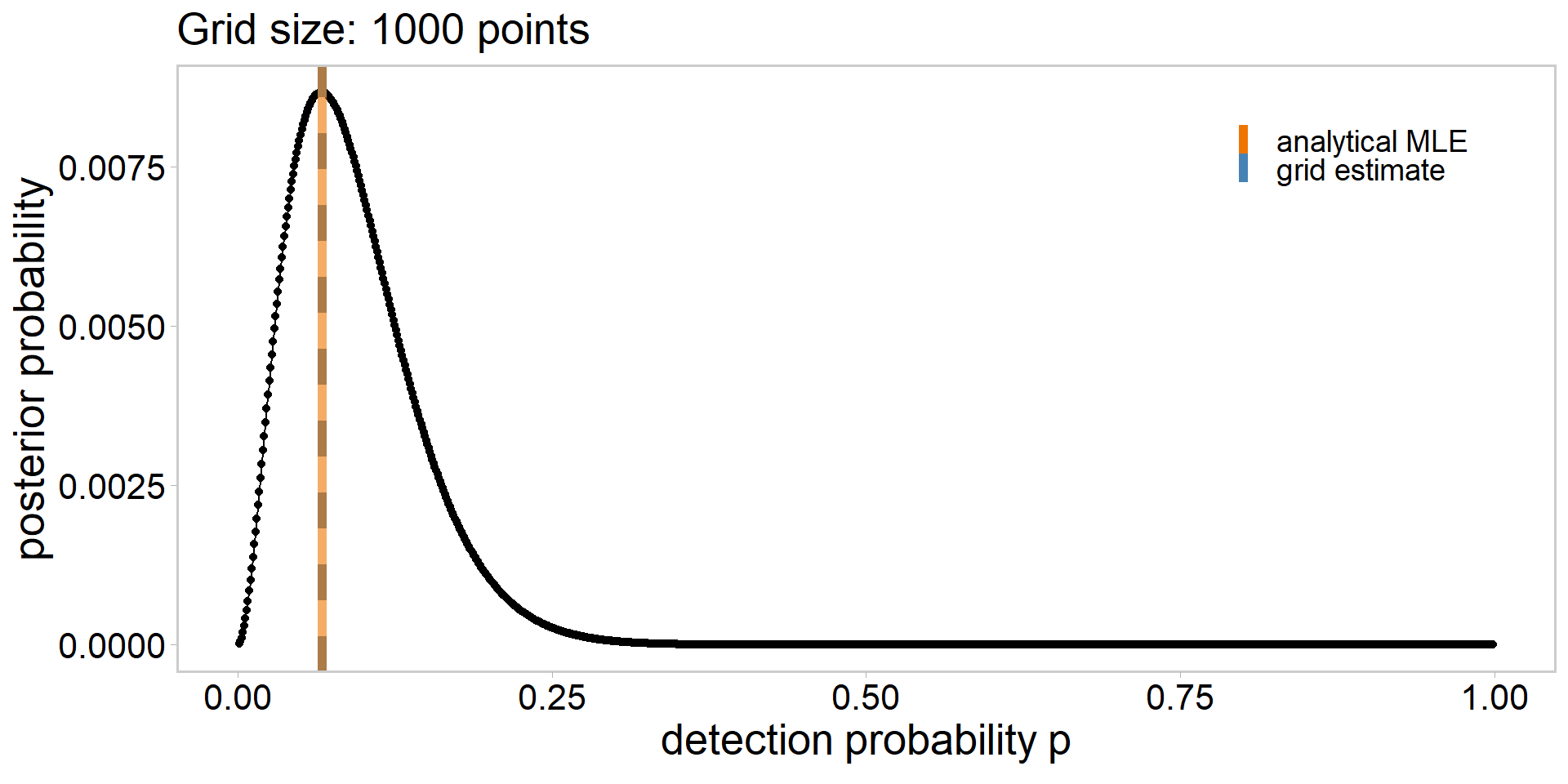
# define a grid
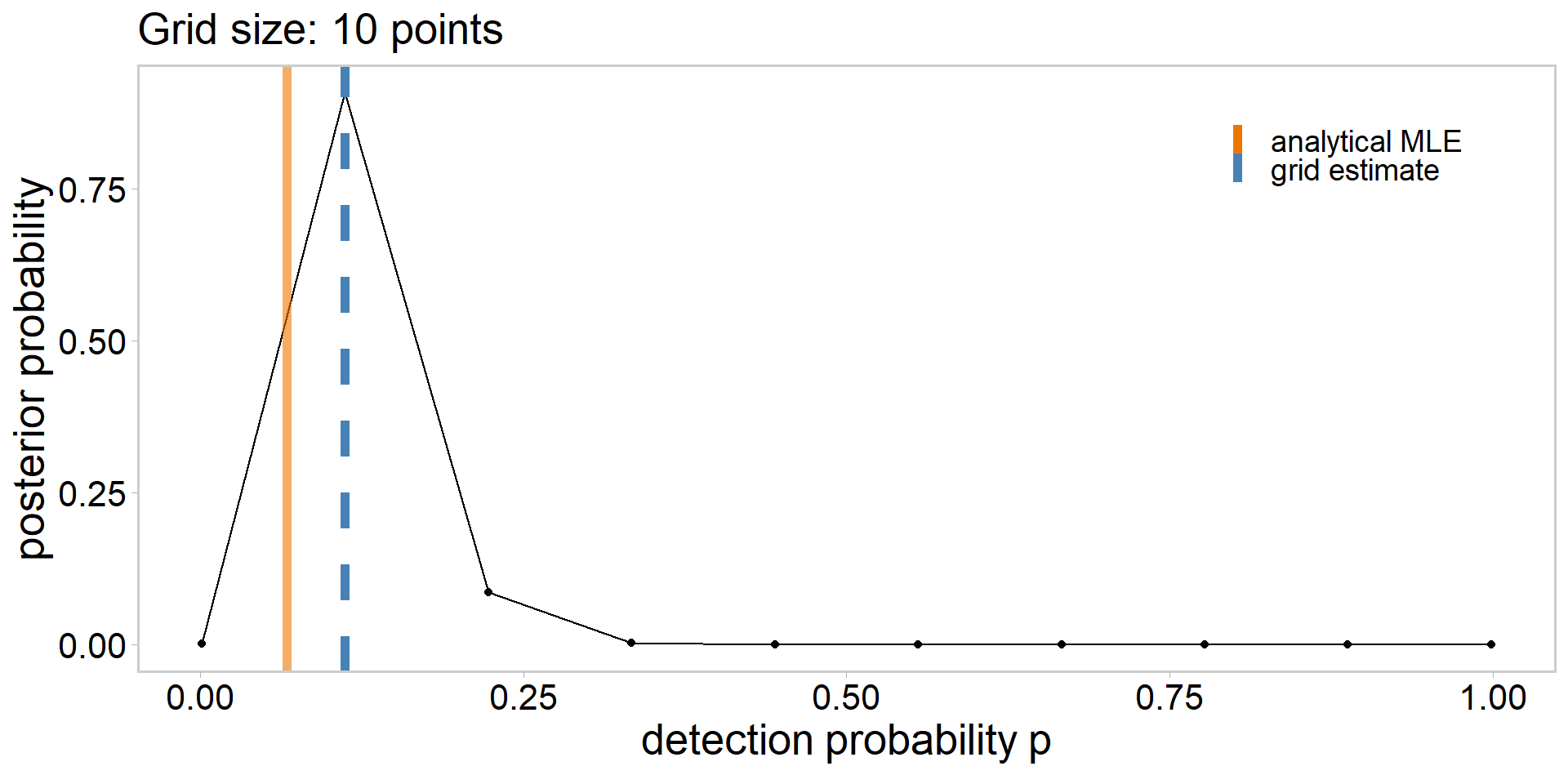
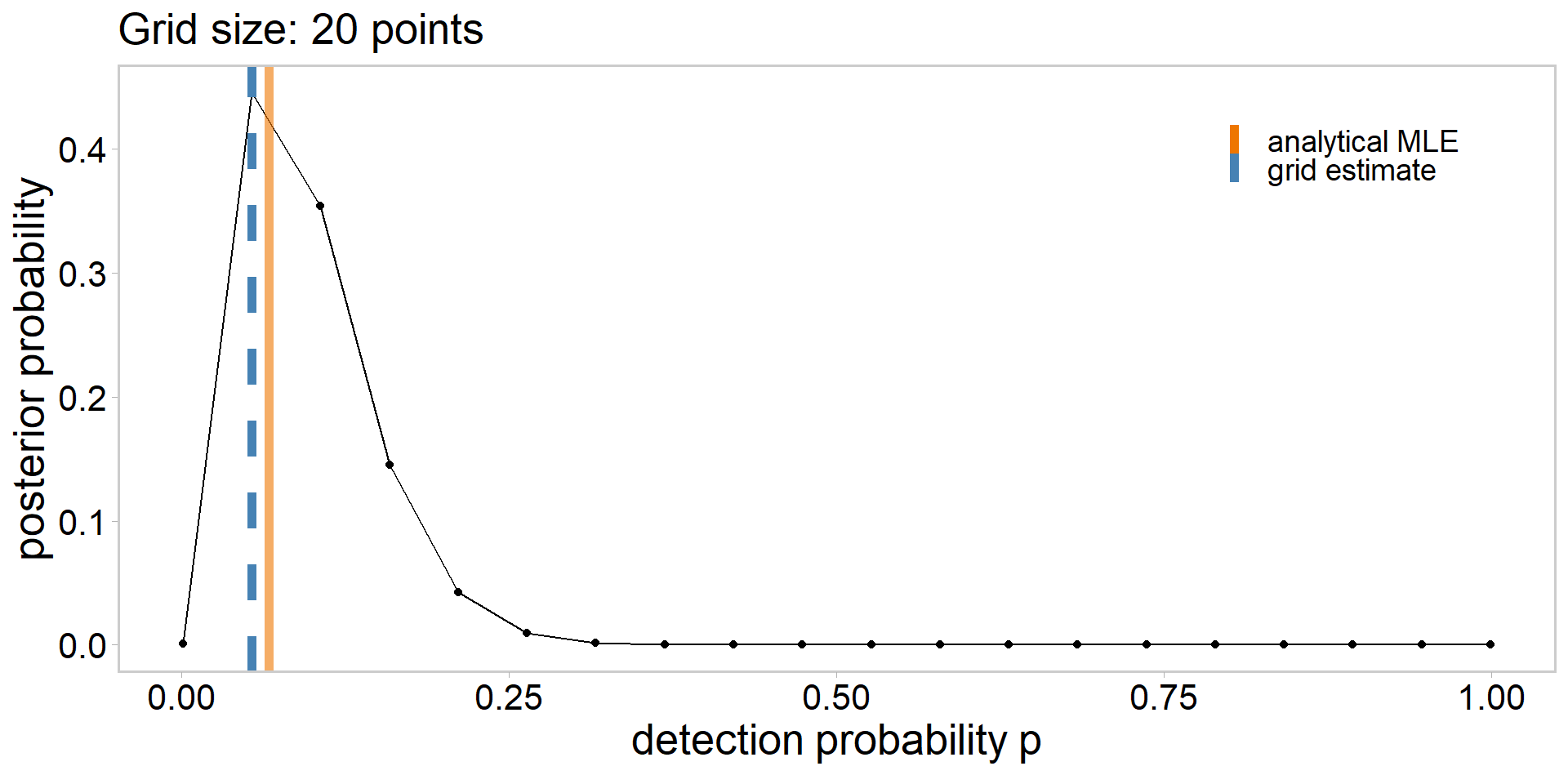
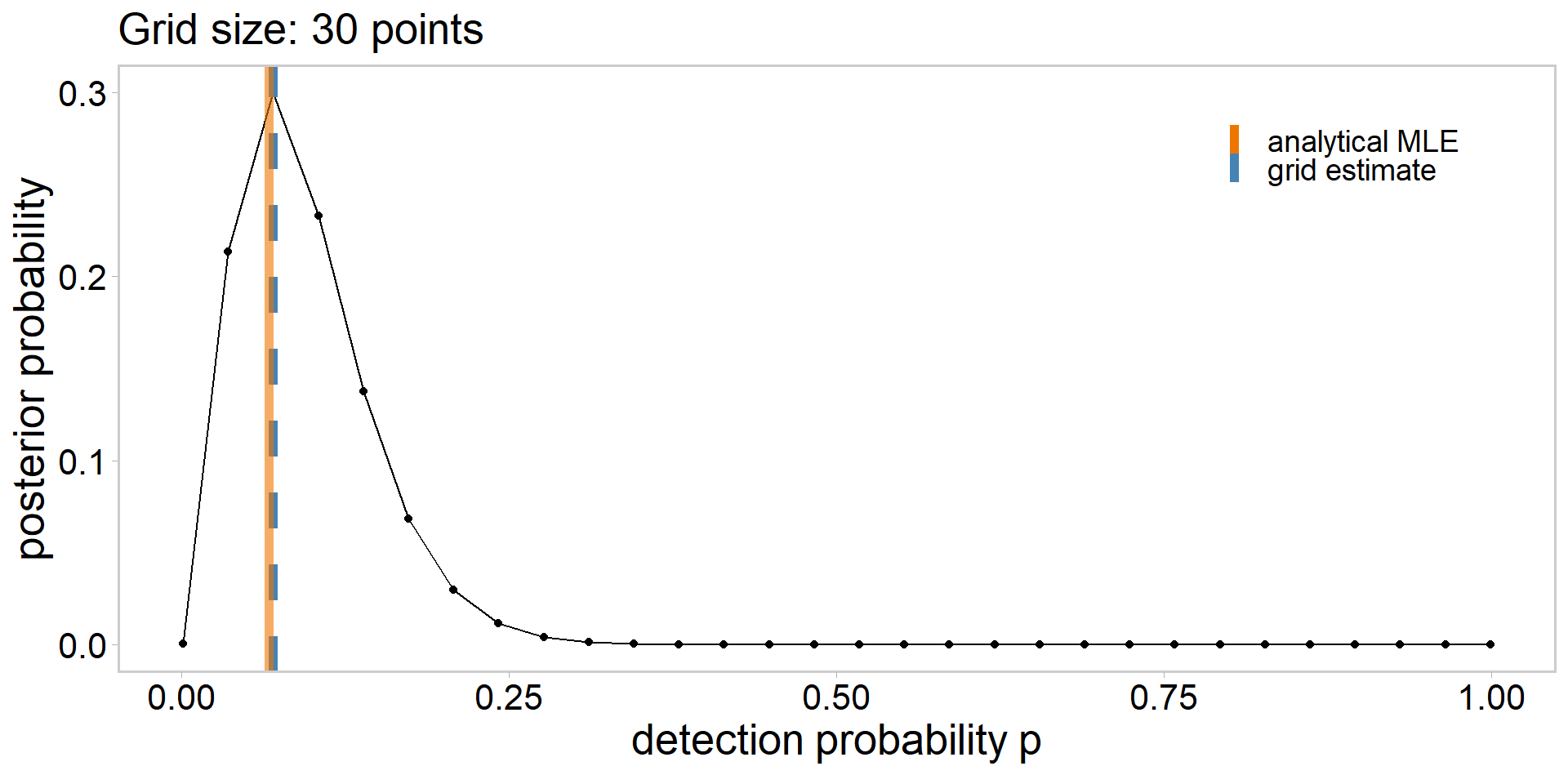
grid_size <- 10
p_grid <- seq(from = 1e-3, to = 0.999, length.out = grid_size)
# calculate likelihood
lik <- dbinom(sum(y), n, p_grid)
lik # likelihood of having observed the data | each p_grid value [1] 4.229830e-04 1.964028e-01 1.859940e-02 5.603740e-04 6.085257e-06
[6] 1.860800e-08 8.682232e-12 1.445413e-16 7.970008e-25 4.341304e-82