1-05: Functions Introduction
0.1 Changes…
1 Purpose
Explain how functions works and how to use the Help tab in RStudio. The presentation of functions in R is inconsistent but it is very useful to understand how to read a function help page.
1.1 Files
The script for the lesson is here
- Save the linked script file to the scripts folder in your Class Project
- All the code in this lesson is sequentially placed in the script
The data used in this lesson, twoWeekWeatherData.csv
- Save the linked data file to the data folder in your Class Project
2 Functions
In the last lesson we used two functions in R:
read.csv(): open up a CSV file and read in the data
seq(): create a configurable sequence of numbers
note: we usually put ( ) after the name of a function to differentiate it from a variable
Functions are standalone scripts designed to do some repeatable operations. A function almost always takes inputs (i.e., arguments) and sends a response (i.e., return value).
In the case of read.csv() from last lesson:
# path to data file (from project folder)
weatherData = read.csv(file="data/twoWeekWeatherData.csv",
sep=",", # values separated by comma
header=TRUE); # there is a header rowThe inputs (arguments) are the values for file, sep, and header.
The response (return value) is the data from the CSV file and gets saved to the variable weatherData.
In the case of seq():
seq1 = seq(from=1, to=10, by=3);The inputs (arguments) are the values for from, to, and by.
The response (return value) is the sequence of numbers that gets saved to the variable seq1.
3 Arguments
In programming, the values you pass into a function (e.g., file, sep, and header) are called arguments of the function. Arguments are added to the function call to tweak the operation of the function – arguments act like the knobs and dials for a function.
Note: The majority of the programming world uses the term parameters instead of arguments. I tend to like parameters better as I think it better indicates how these are properties of the function. But, R is a programming language for statisticians and arguments is a mathematical term, so R chose to go with arguments. Just note you will occasionally see the term parameters, and it means the same thing as arguments.
4 A simpler function: median()
Let’s take a step back and look at a simpler function, median(), which takes a vector and finds the median value in the vector.
To use median() we need a vector with numeric values in it, which we will call vec1:
vec1 = c(3,4,5,6,21,45,61);And then we pass vec1 in as an argument to the median() function:
medianVal1 = median(vec1);Trap: Forgetting to pass in a vector to median
And the Environment shows the return value, saved to medianVal1, is 6
medianVal1: 64.1 Help for median()
Now let’s use RStudio’s Help tab to go behind-the-scenes of median(). RStudio Help tab is a useful resource for finding out more information on functions. The information that appears in Help is from the latest R documentation. If you do an internet search, you will often get older documentation. Note: the Help window is using the website https://search.r-project.org/.
If we type median in the search bar we get this:

The Usage part of the Help page shows that median() has two arguments:
x: the vector that you want to find the median for
na.rm: a TRUE/FALSE value that determines how to handle NA values in the vector. NA means Not Available and usually indicates a problem with the data.
Note: the ( … ) can be ignored – it is R indicating that this function can be expanded with more arguments
4.2 Skipping argument names
When we called median(), we did not include any argument names:
medianVal1 = median(vec1);However, since x is the first argument in median(), it is assumed that the first value is meant to be for x.
This is an equivalent call to median() that more explicitly says that the vec1 is the value for x:
medianVal1b = median(x=vec1);And the return value will be the same:
medianVal1: 6
medianVal1b: 64.3 Default argument values
The other argument in median(), na.rm, has a default value of FALSE . This means that you do not have to supply a value for the argument na.rm when you call the function. You only need to supply a value if you want to change na.rm to something other than the default, FALSE (e.g., TRUE).
Our first example did not have an NA values, let’s create a vector with an NA value in it:
vec2 = c(3,4,5,NA,6,21,45,61);And then use that as an argument for median():
medianVal2 = median(x=vec2);If there is an NA value in a vector then, for most mathematical functions in R, the return value will be NA. The median of a vector with NA values is always NA_real_. NA_real_ means that median cannot be determined but would be a real number if it could be determined.
medianVal2: NA_real_4.4 Dealing with the NA
na.rm is the argument that determines whether NA values are removed from the vector – and the default value is FALSE, meaning NA values are not removed.
Let’s set na.rm to TRUE so the NA values are removed before finding the median:
medianVal2b = median(x=vec2, na.rm=TRUE);Now the NA in the vector is ignored and we get the same median value as before:
medianVal2b: 64.5 Alternate ways to call median()
Argument names (e.g., x and na.rm) are not needed if you put values in the correct order. All of these calls functionally do the same thing as the above median() call:
medianVal2c = median(vec2, TRUE);
medianVal2d = median(vec2, na.rm=TRUE);
medianVal2e = median(na.rm=TRUE, x=vec2); # can reverse arguments if you name them
medianVal2f = median(x=vec2, TRUE);My general recommendation is to always use argument names beyond the first agument (which is usually reserve for the data) when calling a function. This makes your code easier to read and you don’t have to worry about the order of the arguments. This is especially important when you are dealing with more complicated functions that have lots of arguments (e.g., plotting functions).
5 Help seq()
Unfortunately, the Help for functions can contain many abstractions and extraneous information. Throughout this course, we will open up the Help menu for new functions to get you more familiar with the abstractions.
Let’s look at the function seq():
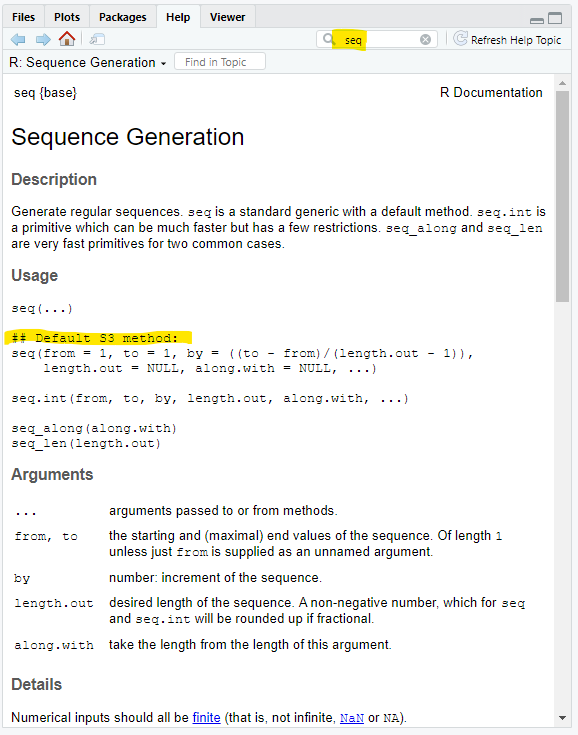
When you see Default S3 Method, that is most likely the version of the function you care about. S3 means S version 3, S is a programming language developed in the 1970s and was the predecessor to R (yeah, that sounds a bit backward…). The other object types you might see are S4, RC (very rare), and R6.
5.1 The arguments and defaults
seq() has 5 arguments: from, to, by, length.out, and along.with. We are going to ignore along.with, which is not needed nor very useful.
The descriptions of the arguments in Figure 3 are helpful but the default argument values for seq() are misinformative:
## Default S3 method
seq(from = 1, to = 1, by = ((to - from)/(length.out - 1)),
length.out = NULL, along.with = NULL, ...)seq() essentially is solving for this equation:
\[ by=\frac{\text{ to }-\text{ from }}{\text{ length.out }-1} \]
There are four variables in the equation and you have to give exactly three out of four of them as arguments. seq() will rearrange the formula, calculate the unknown fourth variable, and create the sequence.
note: every argument in seq() has a default value, meaning you can call seq() without any arguments. The result is not useful, though – but worth a try.
5.2 seq() examples
If you set from, to, and by as we did for seq1, seq() will calculate length.out (the number of values in the sequence)
seq1 = seq(from=1, to=10, by=3); # will have 4 valuesand give you the sequence:
seq1: num[1:5] 1 4 7 10length.out allows you to set the number of values in the sequence. You can also set length.out to a value, but then you can only use 2 of these 3 arguments: from, to, and by.
An example of using length.out:
seq2 = seq(from=1, to=10, length.out=5);Then you get 5 evenly spaced number starting with 1 and ending with 10
seq2: num[1:5] 1 3.25 5.5 7.75 106 read.csv() expanded
When you go to the Help page for read.csv(), you are taken to the Help page for read.table(). This is because read.csv() is a version of the function read.table(). In other words, read.csv() is read.table() with different default values for the arguments. The different argument values are underlined in Figure 4.
For instance, the default value for sep (the separator between data) is a space in read.table() and a comma in read.csv(). read.csv() uses all the argument from read.table() and changes some of argument value (the arguments underlined below).
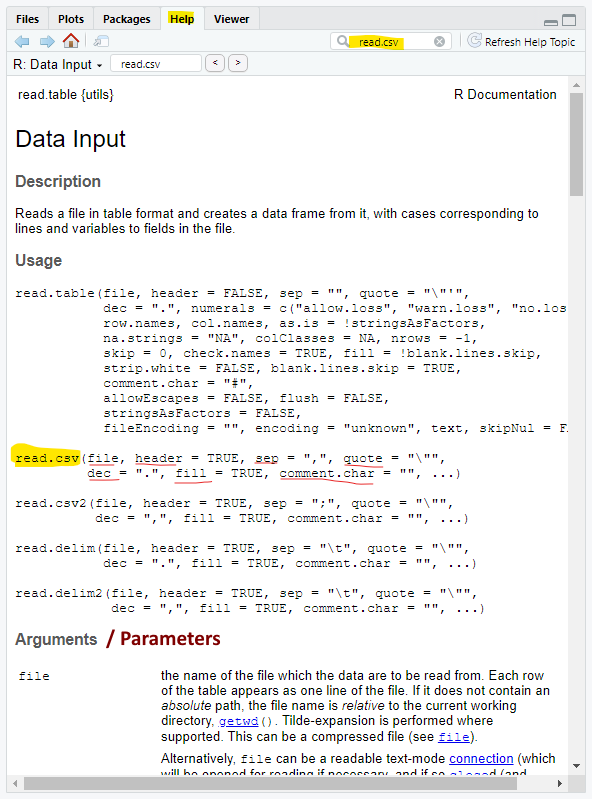
6.1 read.csv arguments
When you call read.csv(), you can use any of the argument from read.table(). For instance allowEscapes and stringsAsFactors are arguments listed only in read.table(), but they can be used in read.csv().
And there are a lot of arguments in read.table(). The majority of these arguments rarely need to be changed from their default value.
Most of the arguments in read.table() look cryptic and you will probably never have to use them. A couple that are easier to understand:
decimal: the character that represents a decimal in the decimal number. You might need to change this to a comma ( , ) if you get data from Europe
comment.char: the character after which everything on the line is a comment (i.e., not data)
- notice that read.csv() has no comment.char whereas read.table() uses ( # ) as the default comment.char
6.2 Required arguments and default arguments
file is an argument that is not set equal to anything in read.table() or read.csv(). This is because file must be supplied by the user –file is the only required argument in either of these functions. This should make sense as there is no point in calling read.table() / read.csv() without a data file.
Most other argument in read.table() / read.csv() has a default value (except row.names and col.names – there is a question about this in the application). This means you can execute the function without using any argument except file.
In fact, the following code will produce the same results as the earlier read.csv() call in Figure 1:
weatherData2 = read.csv(file = "data/twoWeekWeatherData.csv");If you look in the Environment tab the values for weatherData and weatherData2 are exactly the same.
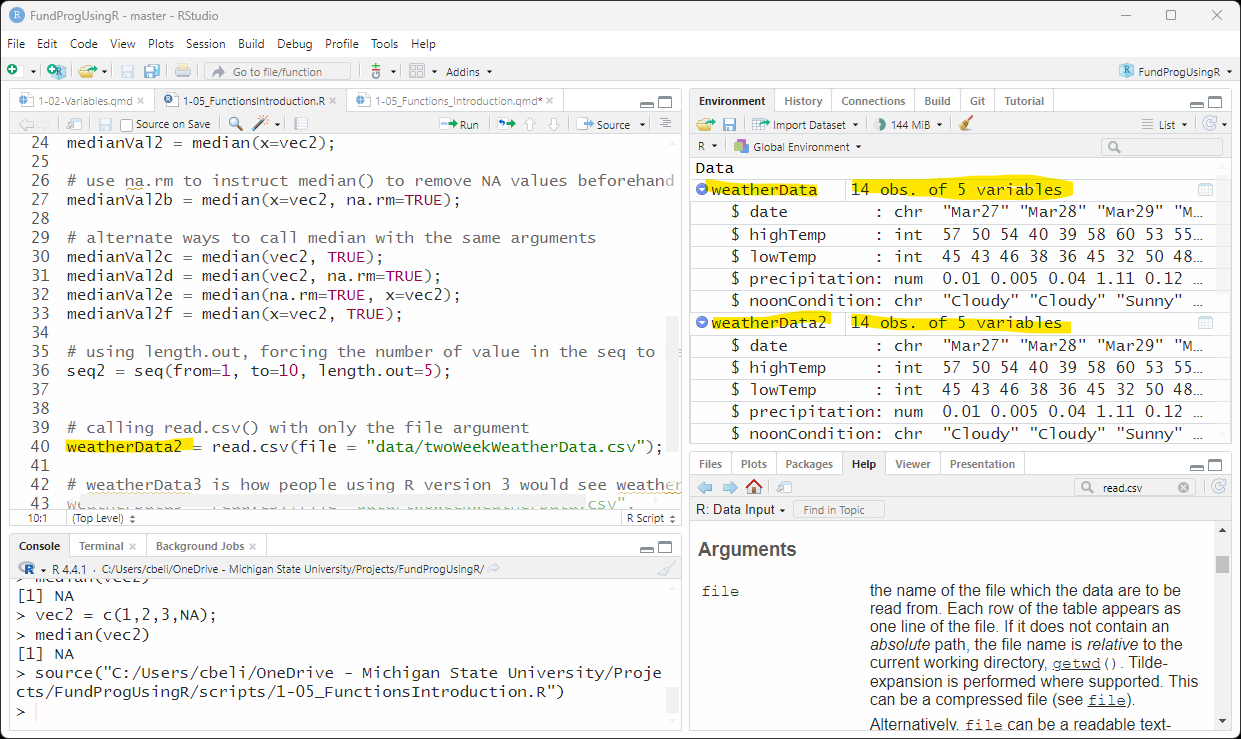
6.3 The other arguments (aside from file)
In my original call to read.csv(), the arguments sep=“,” and header=TRUE are functionally not changing anything because I have them set to the same value as their default. But, these arguments are changed often enough that it is nice to have them quickly at hand when copying read.csv() from one script to another.
weatherData = read.csv(file="data/twoWeekWeatherData.csv",
sep=",",
header=TRUE);7 Application
1. Create this sequence: 13, 9, 5, 1, -3, -7, -11 using seq().
- Come up with two more ways (three in all) to produce the same sequence using seq(), but changing the arguments (from, by, to, length.out).
2. Explain, in comments, why you get an error if you try to use all four arguments (from, by, to, length.out)
3. In comments answer: In read.csv(), what are the default values for row.names, na.strings, fill , and comment.char? Note: some are defined in read.table(), some are defined in read.csv().
4. Find the log (base 5) of this vector: c(0.04, 0.2, 25, 125) using the log() function
Use the Help tab to find out more about the log() function
The answer is: -2, -1, 2, 3 (so you can check)
note: the default value for base is exp(1) = e1 = e = 2.71828… (i.e., the natural logarithm)
5. Properly read in this CSV file of the same two week weather data but:
commas are used in place of decimals in precipitation column
spaces are used to separate variables (instead of commas)
Save the data to a dataframe name WD_Comma
6. Call the function factor() twice, passing in (1) the column date and (2) the column noonCondition as the argument and answer the following in comments:
How many factors levels does the noonCondition column have?
How many factors levels does the date column have?
What is a factor level?
Save the script as app1-05.r in your scripts folder and email your Project Folder to Charlie Belinsky at belinsky@msu.edu.
Instructions for zipping the Project Folder are here.
If you have any questions regarding this application, feel free to email them to Charlie Belinsky at belinsky@msu.edu.
7.1 Questions to answer
Answer the following in comments inside your application script:
What was your level of comfort with the lesson/application?
What areas of the lesson/application confused or still confuses you?
What are some things you would like to know more about that is related to, but not covered in, this lesson?
8 Trap: Forgetting to pass in a vector to median
If you want to find the median of the vector: c(3,4,6,2,7,10), you have two choices:
Save the vector to a variable and use the variable as an argument to median():
> vec6 = c(3,4,6,2,7,10)
> median(vec6)
[1] 5Use the vector directly as an argument to median():
> median(c(3,4,6,2,7,10))
[1] 5Both produce the correct answer, 3.5.
A mistake people often make is to forget to put the numbers into a vector by using c(), they do this instead:
> median(3,4,6,2,7,10)
[1] 3Is this case, you are calling median() with 6 separate arguments: 3, 4, 6, 2, 7, and 10. median() only takes 1 arguments so it ignores the last five number and takes the median of 3, which is 3.
When you put the six numbers into c(), you are saying that these numbers are all grouped together into one vector, and median will take in the whole vector as the argument.